
DivBrowse is a software for interactive visualization and analysis of the diversity of genomic variants. It uses VCF and GFF3 files as data input and consists of a web server written in Python and a GUI written in Javascript. It is available as a Python package on PyPI.org.
Demo instances
| Species | # Variants | # Genotypes | Description | URLs / DOIs of dataset | DivBrowse instance |
|---|---|---|---|---|---|
| Homo sapiens | 73,159,510 | 2548 | Biallelic SNVs called from 2,548 human samples across 26 populations from the 1000 Genomes Project, called directly against GRCh38 | VCF: https://www.ebi.ac.uk/ena/browser/view/PRJEB30460 GFF3: https://www.ensembl.org/Homo_sapiens/Info/Index |
Open |
| Mus musculus | 78,772,544 | 36 | Sanger Institute Mouse Genomes Project v5: SNP calls from version 5 of the Mouse Genome Project at the Wellcome Trust Sanger Institute. Specifically, this project describes the variants of 36 mouse strains aligned against the reference mouse genome sequence GRCm38. | VCF: https://www.ebi.ac.uk/ena/browser/view/PRJEB11471 GFF3: https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/ |
Open |
| Hordeum vulgare | 1,052,403 | 22626 | Unimputed SNP variants for 22626 Hordeum vulgare genotypes called against the reference genome "Morex V3" | VCF: http://doi.org/10.5447/ipk/2021/3 GFF3: http://doi.org/10.5447/ipk/2021/3 |
Open |
| Hordeum vulgare | 223,387,147 | 300 | Unimputed SNP variants for 300 Hordeum vulgare genotypes called against the reference genome "Morex V2" | VCF: https://doi.org/10.5447/ipk/2020/24 GFF3: https://doi.org/10.5447/IPK/2019/8 Paper: https://doi.org/10.1038/s41586-020-2947-8 |
Open |
| Triticum aestivum | 1,628,276 | 8070 | Unimputed SNP variants for 8070 Triticum aestivum genotypes called against the reference genome "IWGSC Chinese Spring RefSeq v1.0" | VCF: https://www.ebi.ac.uk/ena/browser/view/PRJEB52759 GFF3: https://ftp.ensemblgenomes.ebi.ac.uk/pub/plants/release-55/gff3/triticum_aestivum/ Paper: https://doi.org/10.1038/s41588-022-01189-7 |
Open |
| Triticum aestivum | 213,804,916 | 768 | Unimputed SNP variants for 768 Triticum aestivum genotypes called against the reference genome "IWGSC Chinese Spring RefSeq v1.0" | VCF: https://www.ebi.ac.uk/ena/browser/view/PRJEB52759 GFF3: https://ftp.ensemblgenomes.ebi.ac.uk/pub/plants/release-55/gff3/triticum_aestivum/ Paper: https://doi.org/10.1038/s41588-022-01189-7 |
Open |
Key facts
- Uses established bioinformatics file formats like VCF and GFF3 for data input
- Can handle very large VCF files up to 1 TB (gzip compressed)
- Configureable via YAML config file
- Filtering of variants by e.g. MAF, heterozygosity, QUAL-values
- Sorting of genotypes by name (alphabetical) and phylogenetic distance
- Counting of variants for each gene and their exons
- Built-in interactive analysis features like PCA, UMAP
- Export of VCF files
- Export of GFF3 files
Features
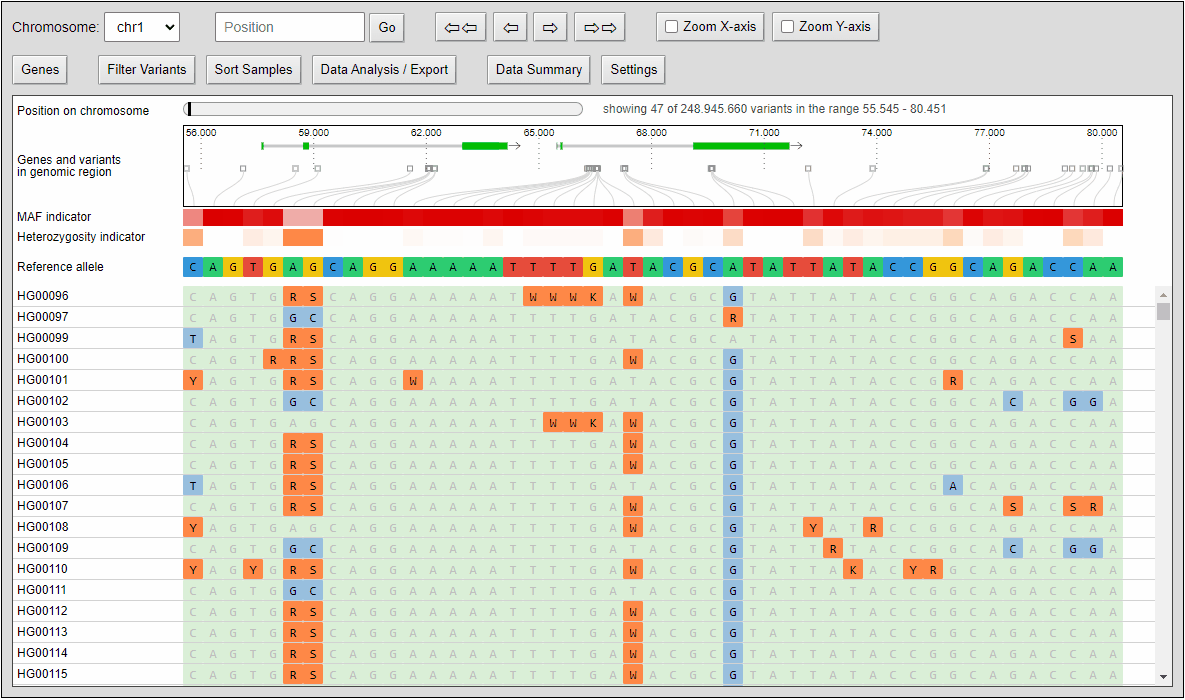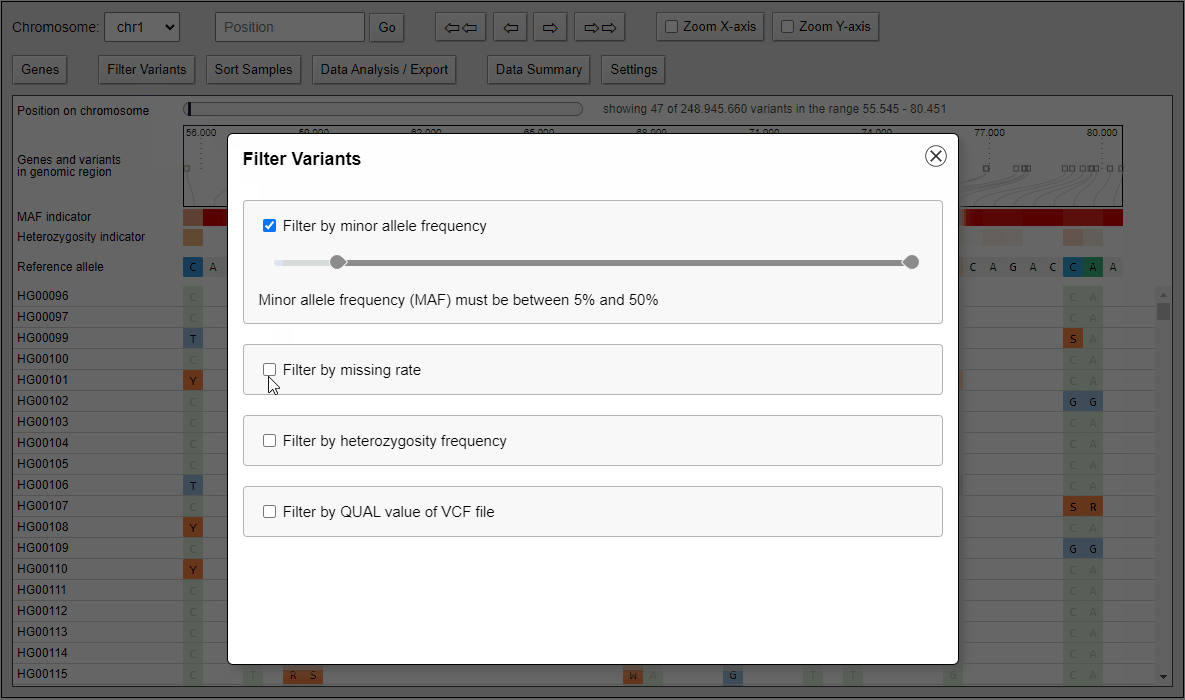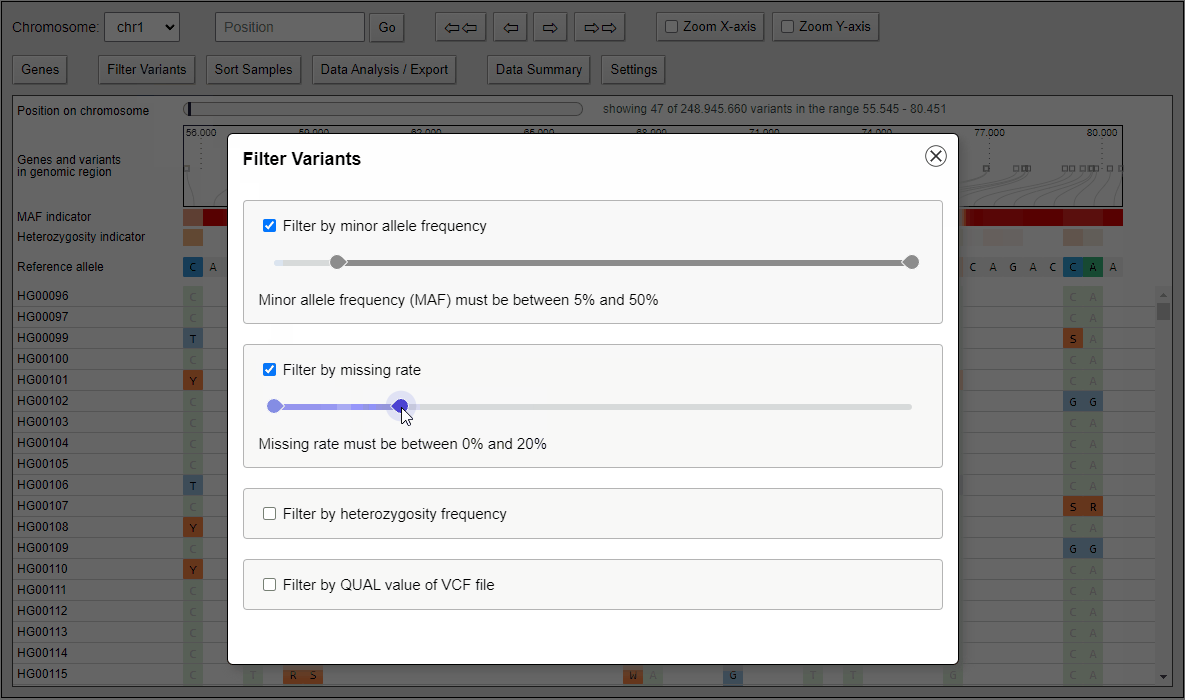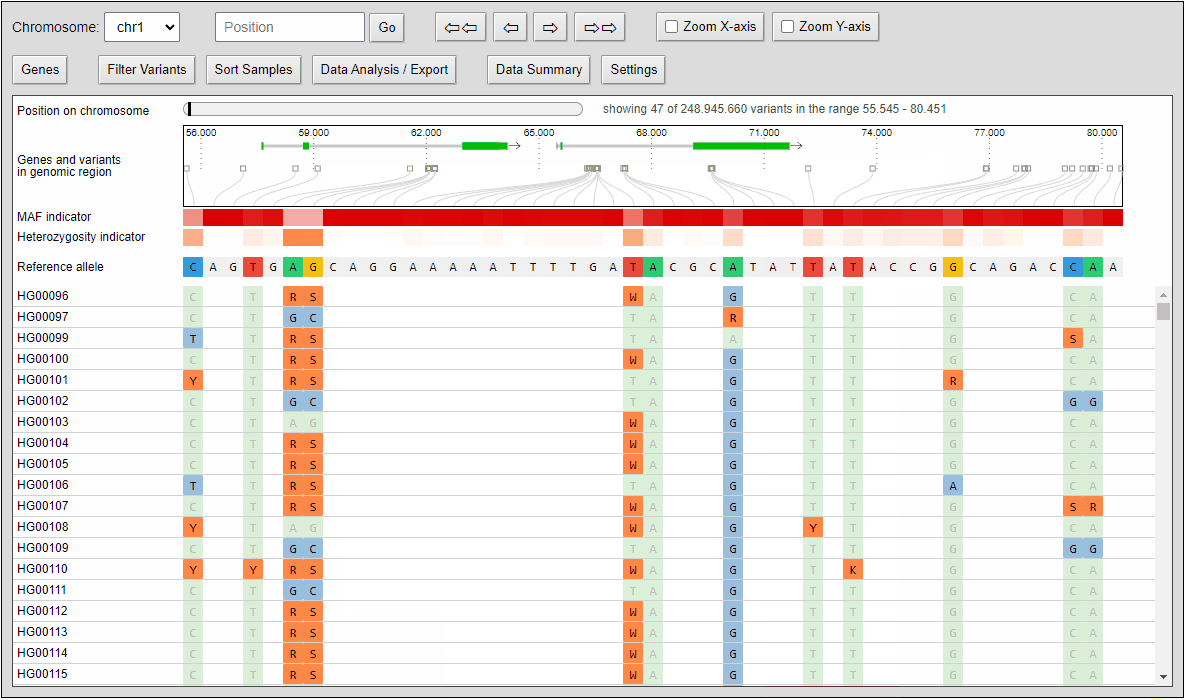
Filtering of variants
Quickly filter variants on calculated statistical measures like minor allele frequency, heterozygosity frequency or even QUAL values that come directly with the VCF file. Filter settings can also applied to data analysis features like the built-in principal component analysis and to the VCF export.
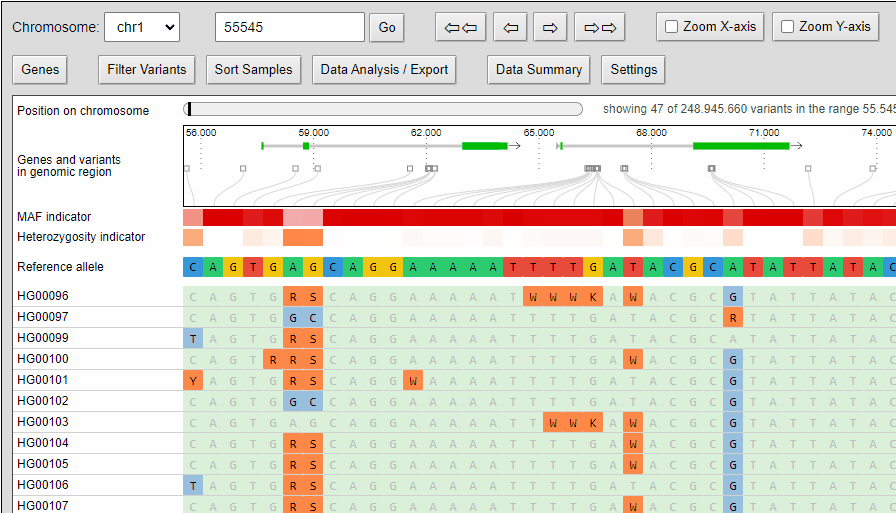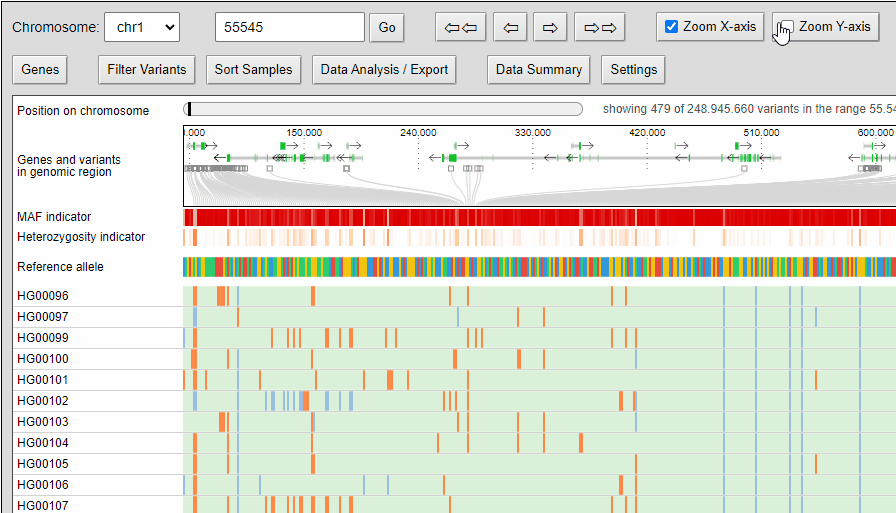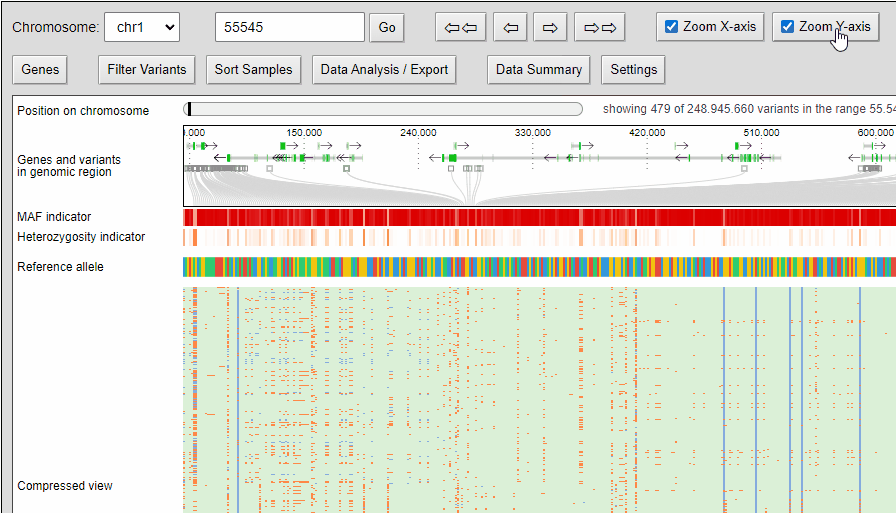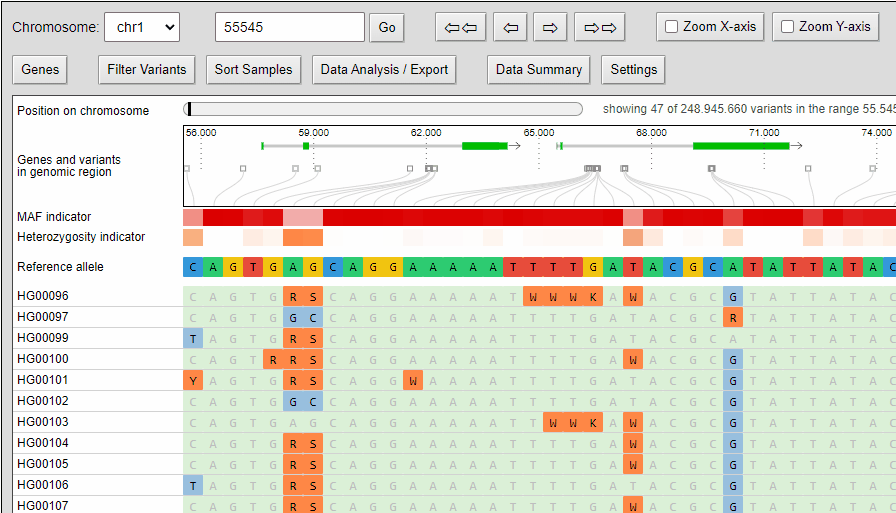
Zooming
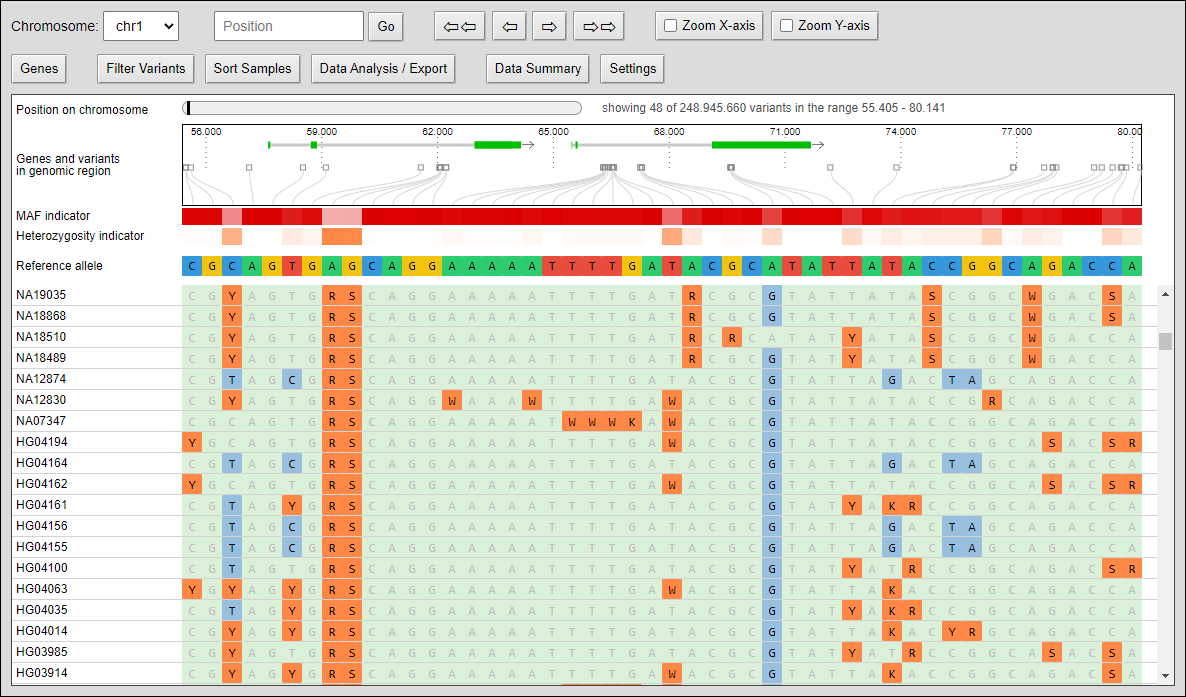
The compressed view shows each genotype of the VCF as a one pixel high line. If your diversity panel contains of many genotypes, more genotypes can be visualized at once.
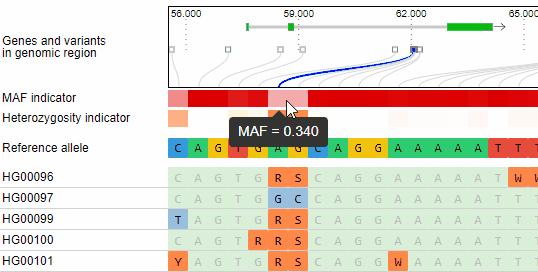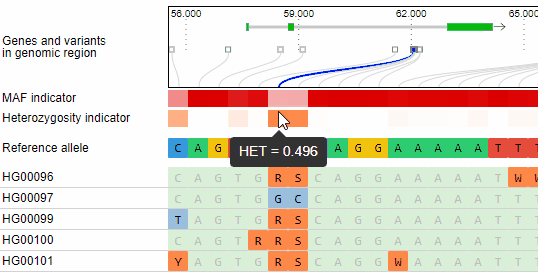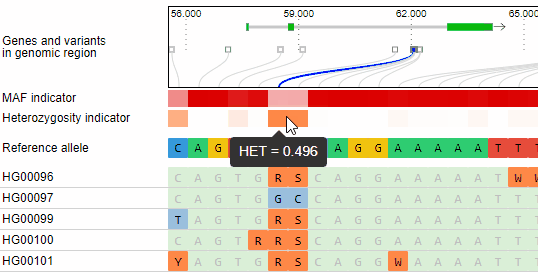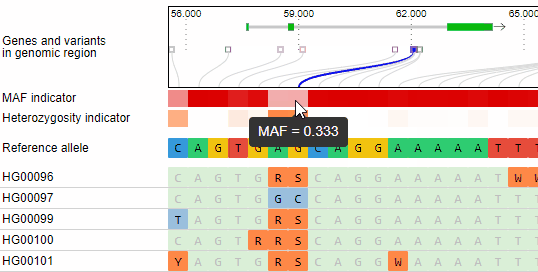
Minor allele frequency and heterozygosity calculation
The minor allele frequency and percentage of heterozygous calls of each variant is calculated ad-hoc for the given set of genotypes. The MAF is visualized in a horizontal heat map. The concrete MAF value can be obtained by moving the mouse cursor over the variant.
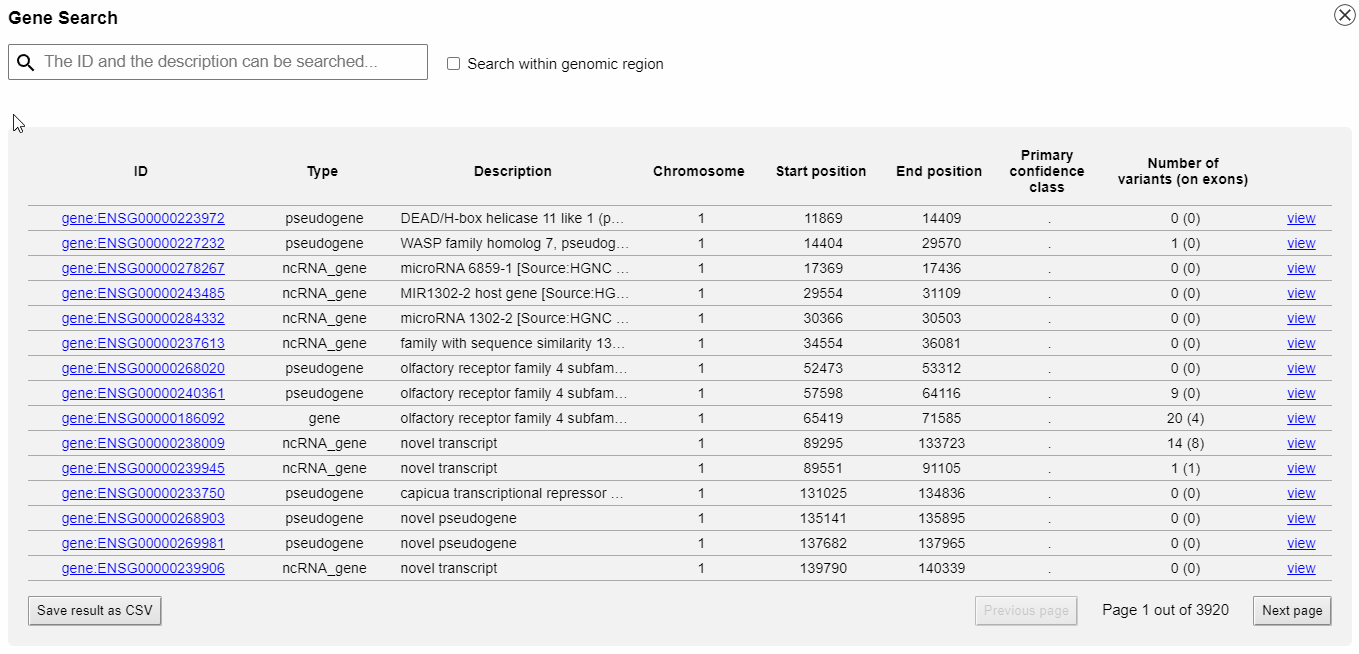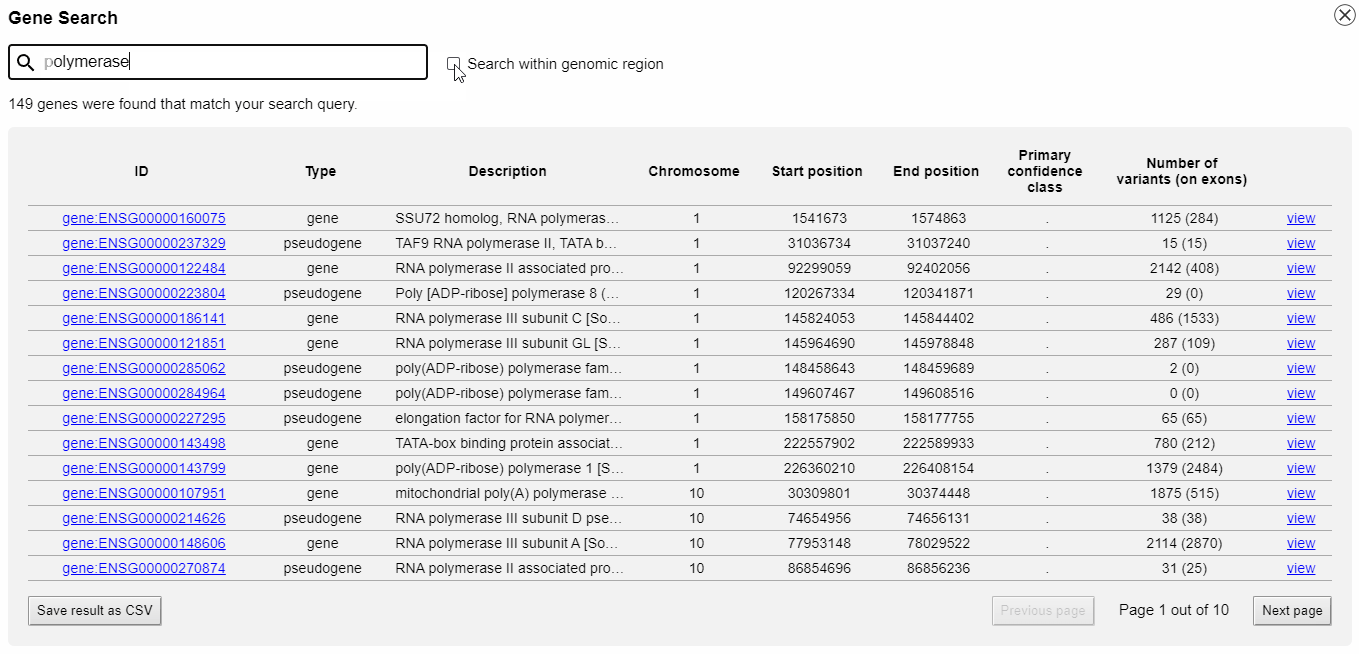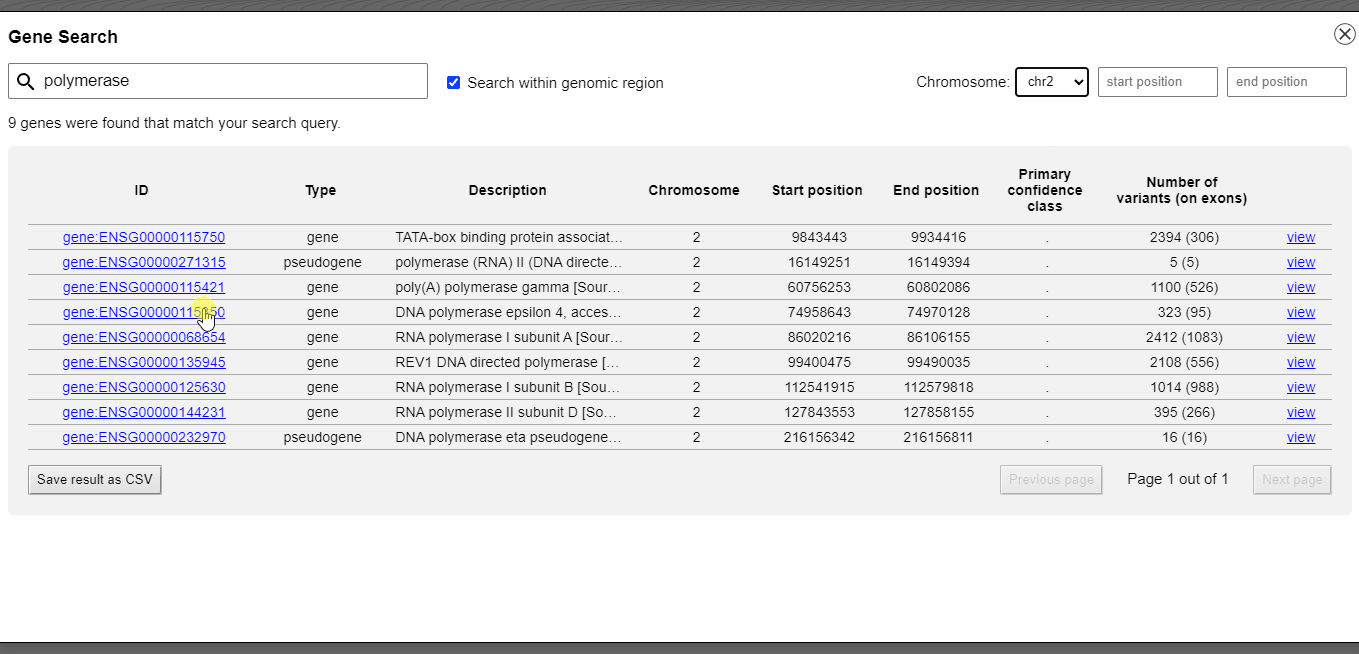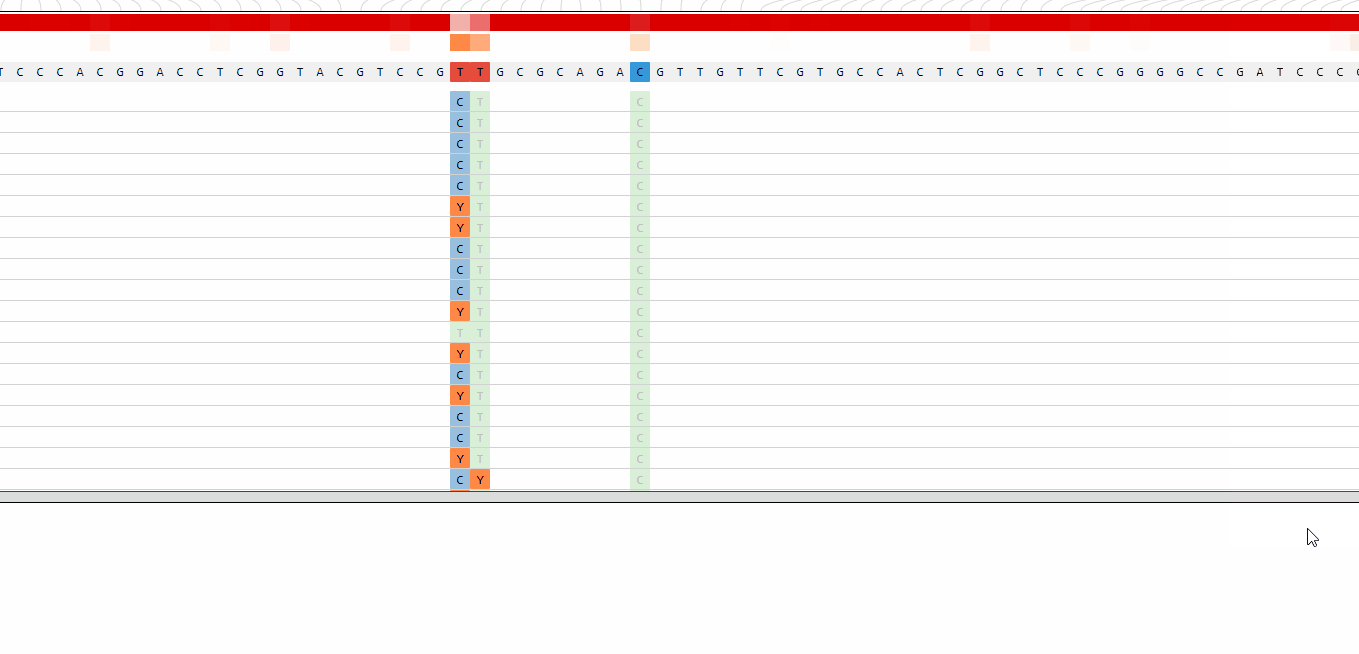
Gene catalogue and search
With the integrated gene catalogue you can browse and search all genes that are provided by your GFF3 file. The search can be restricted to a specific genomic region. You can directly jump to a gene without typing in or copy-and-paste genomic coordinates.

Sorting of genotypes
The list of genotypes can be sorted alphabetically or by the phylogenetic distance to the reference genome.
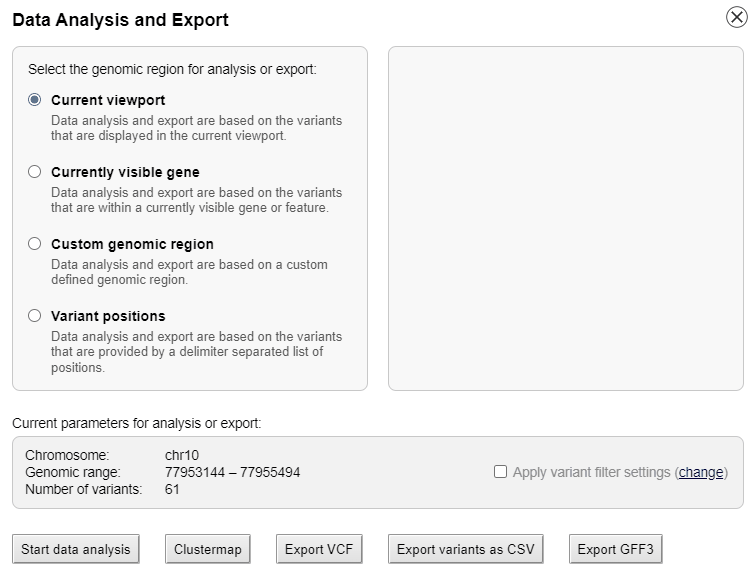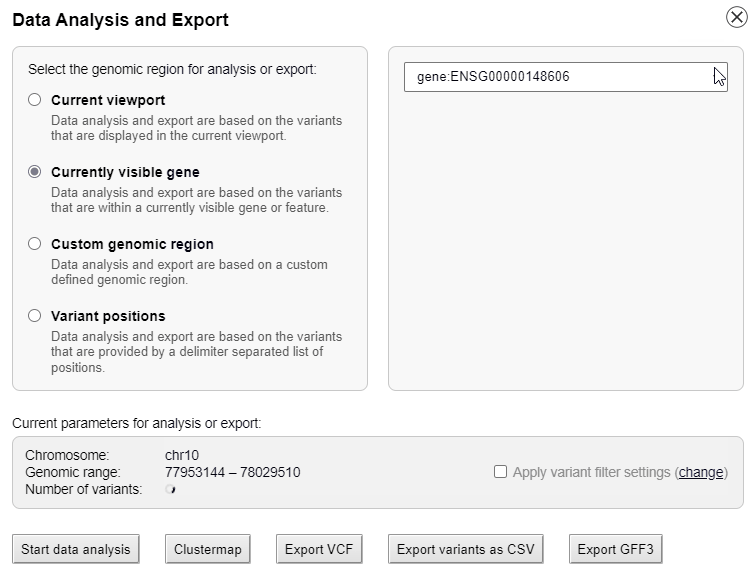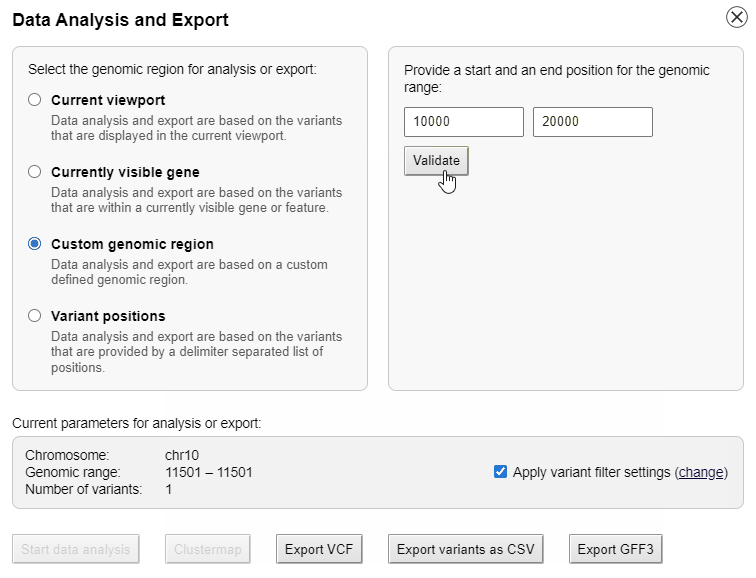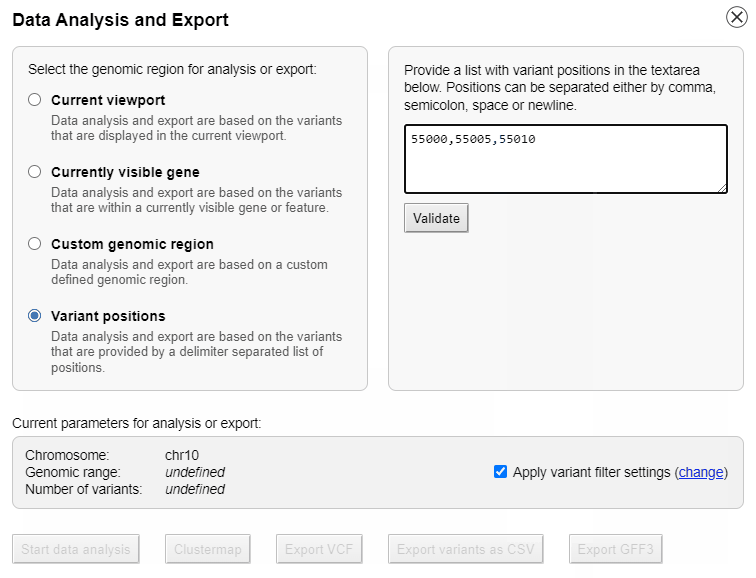
Interactive data analysis
The list of genotypes can be sorted alphabetically or by the phylogenetic distance to the reference genome.
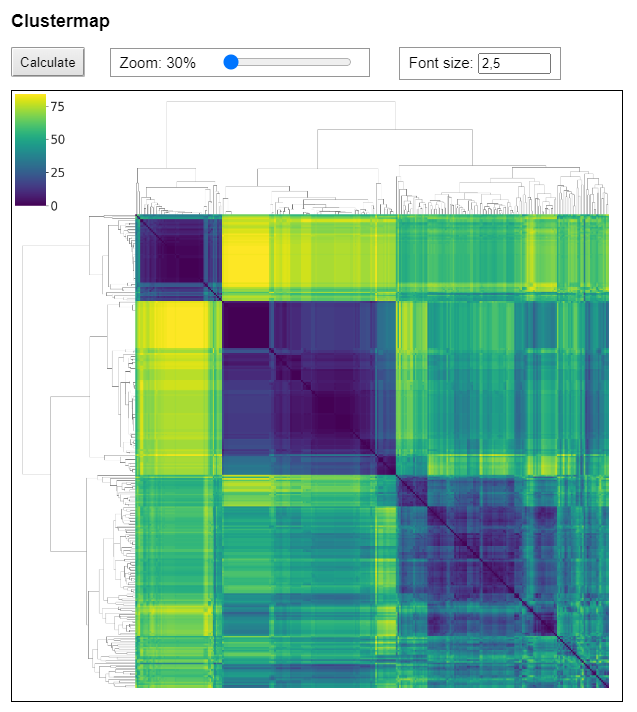
Hierarchically-clustered heatmaps of distance matrices
Hierarchically-clustered heatmaps of distance matrices can be created based on custom genomic ranges of interest, e.g. for single genes, exons or even larger genomic regions.